Few days ago, I had to quickly create 2 plots to ask some questions in a PhD quiz. This is the code.
Required
Load required packages (tidyverse and cowplot):
library(tidyverse)
library(cowplot)Data
Make some random data:
# 5 vectors that will be 4 groups:
raw <- data.frame(
ctrl = rnorm(15,mean=2,sd=.7),
mela = rnorm(15,mean=2.8,sd=.7),
melb = rnorm(15,mean=3.2,sd=.7),
melc = rnorm(15,mean=3.4,sd=.7),
Group = c('treatment')
)Gather columns into rows:
df <- raw %>%
gather(Group,Proliferation,1:4)Reorder groups by ascendent concentration of melatonin, and label:
df$Group <- factor(
df$Group,
levels=c('ctrl','mela','melb','melc'),
labels=c('Control','Mel 0.1nM','Mel 1nM','Mel 10nM')
)
str(df)## 'data.frame': 60 obs. of 2 variables:
## $ Group : Factor w/ 4 levels "Control","Mel 0.1nM",..: 1 1 1 1 1 1 1 1 1 1 ...
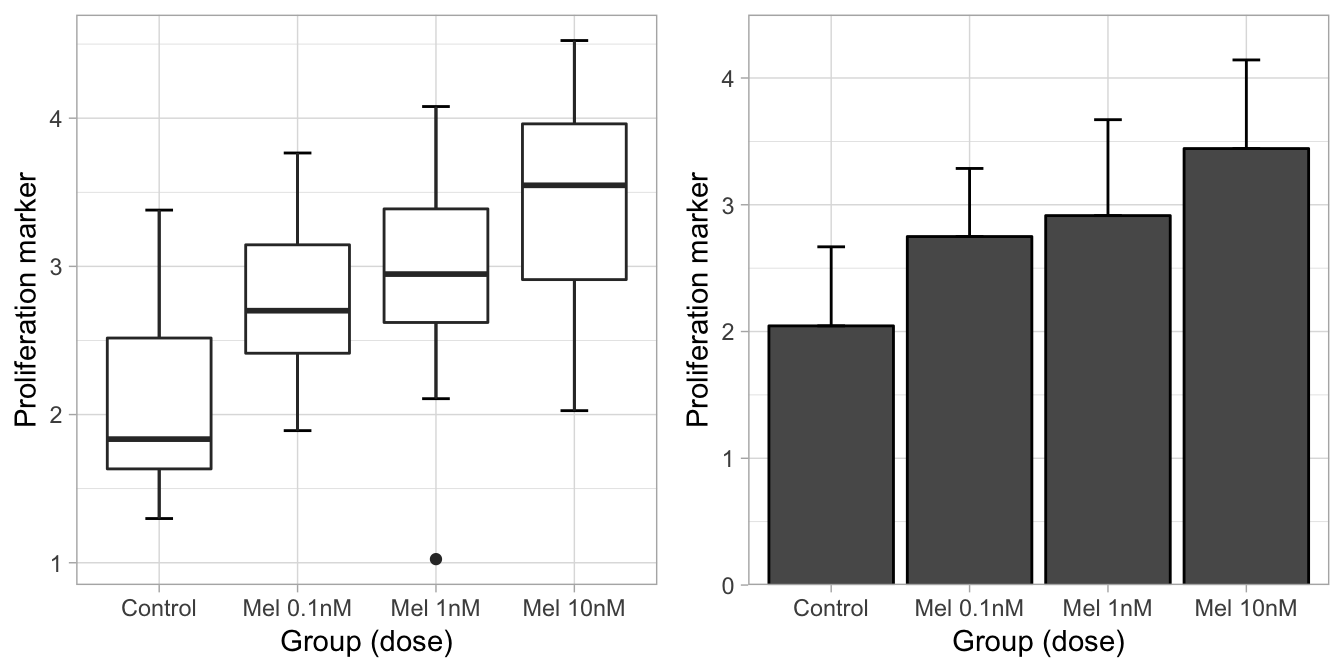
## $ Proliferation: num 1.61 2.82 2.79 3.38 1.87 ...Ok, we have a proliferation vector measured as fluorescence of some surface protein marker, separated into 4 groups: Control, Melatonin 0.1 nM, Melatonin 1 nM, and Melatonin 10 nM.
Plot
Let’s plot!
Make a Box plot:
boxplot <- ggplot(df,aes(x=Group,
y=Proliferation)
)+
stat_boxplot(geom='errorbar',width=.2)+
geom_boxplot()+
scale_x_discrete(name='Group (dose)')+
scale_y_continuous(name='Proliferation marker')+
theme_light()Make a bar plot, with mean and standard deviation bars:
# Summarize (needed for the barplot)
bars <- df %>%
group_by(Group) %>%
summarize(proliferation=mean(Proliferation),
sd=sd(Proliferation)
)
# Plot
barplot <- ggplot(bars,aes(x=Group,
y=proliferation)
)+
stat_summary(fun.y='mean',geom='bar',color='black')+
geom_errorbar(width=.2,aes(ymin=proliferation,
ymax=proliferation+sd))+
scale_x_discrete(name='Group (dose)')+
scale_y_continuous(name='Proliferation marker',
expand=c(0,0),limits=c(0,4.5)
)+
theme_light()Print layout:
ggdraw()+
draw_plot(boxplot,0,0,.5,1)+
draw_plot(barplot,.5,0,.5,1)